Searchlight hyperalignment#
In this Notebook, we will go through how to perform searchlight hyperalignment between brains.
Preparations#
%%capture
%pip install -U neuroboros
import numpy as np
import neuroboros as nb
from scipy.stats import zscore
from scipy.spatial.distance import pdist, cdist, squareform
import matplotlib.pyplot as plt
from joblib import Parallel, delayed
from hyperalignment.procrustes import procrustes
from hyperalignment.ridge import ridge
from hyperalignment.searchlight import searchlight_procrustes, searchlight_ridge
The Life dataset contains fMRI data (a) during watching the Life documentary and (b) during watching videos of various actions of various animals. See Nastase et al. (2017, 2018) for more details.
This example focuses on the movie data (Life documentary).
dset = nb.Life()
sids = dset.subjects
dm1_train = dset.get_data(sids[0], 'life', [1, 2], 'l')
dm1_test = dset.get_data(sids[0], 'life', [3, 4], 'l')
dm2_train = dset.get_data(sids[1], 'life', [1, 2], 'l')
dm2_test = dset.get_data(sids[1], 'life', [3, 4], 'l')
Response hyperalignment#
radius = 20
sls, dists = nb.sls('l', radius, return_dists=True)
xfm_procr = searchlight_procrustes(dm1_train, dm2_train, sls, dists, radius)
xfm_ridge = searchlight_ridge(dm1_train, dm2_train, sls, dists, radius)
pred_procr = dm1_test @ xfm_procr
pred_ridge = dm1_test @ xfm_ridge
isc_orig = np.mean(zscore(dm1_test, axis=0) * zscore(dm2_test, axis=0), axis=0)
isc_procr = np.mean(zscore(pred_procr, axis=0) * zscore(dm2_test, axis=0), axis=0)
isc_ridge = np.mean(zscore(pred_ridge, axis=0) * zscore(dm2_test, axis=0), axis=0)
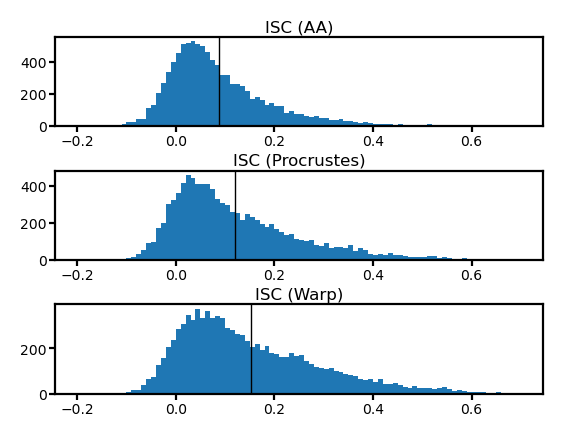
fig, axs = plt.subplots(3, 1, figsize=[_/2.54 for _ in [8, 6]], dpi=200)
for i, (ax, isc) in enumerate(zip(axs, [isc_orig, isc_procr, isc_ridge])):
ax.hist(isc, np.linspace(-0.2, 0.7, 91))
ax.axvline(isc.mean(), color='k', lw=0.5)
ax.tick_params('both', size=2, pad=1, labelsize=5)
title = ['ISC (AA)', 'ISC (Procrustes)', 'ISC (Warp)'][i]
ax.set_title(title, size=6, pad=2)
fig.subplots_adjust(hspace=0.5)
plt.show()
Connectivity hyperalignment#
mapper = nb.mapping('l', 'onavg-ico32', 'onavg-ico8', mask=True)
dm1_ico8 = dm1_train @ mapper
dm2_ico8 = dm2_train @ mapper
conn1 = 1 - cdist(dm1_ico8.T, dm1_train.T, 'correlation')
conn2 = 1 - cdist(dm1_ico8.T, dm2_train.T, 'correlation')
cha_procr = searchlight_procrustes(conn1, conn2, sls, dists, radius)
cha_ridge = searchlight_ridge(conn1, conn2, sls, dists, radius)
pred_procr_cha = dm1_test @ cha_procr
pred_ridge_cha = dm1_test @ cha_ridge
isc_procr_cha = np.mean(zscore(pred_procr_cha, axis=0) * zscore(dm2_test, axis=0), axis=0)
isc_ridge_cha = np.mean(zscore(pred_ridge_cha, axis=0) * zscore(dm2_test, axis=0), axis=0)
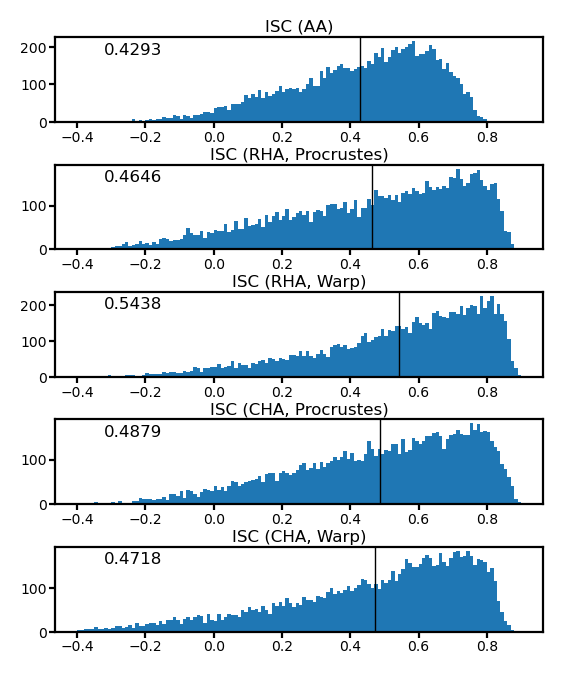
fig, axs = plt.subplots(5, 1, figsize=[_/2.54 for _ in [8, 10]], dpi=200)
for i, (ax, isc) in enumerate(zip(axs, [isc_orig, isc_procr, isc_ridge, isc_procr_cha, isc_ridge_cha])):
ax.hist(isc, np.linspace(-0.2, 0.7, 91))
ax.axvline(isc.mean(), color='k', lw=0.5)
ax.tick_params('both', size=2, pad=1, labelsize=5)
title = ['ISC (AA)', 'ISC (RHA, Procrustes)', 'ISC (RHA, Warp)',
'ISC (CHA, Procrustes)', 'ISC (CHA, Warp)'][i]
ax.set_title(title, size=6, pad=2)
ax.annotate(f'{isc.mean():.4f}', (0.1, 0.8), xycoords='axes fraction', size=6)
fig.subplots_adjust(hspace=0.5)
plt.show()
Connectivity profiles#
nt, nv = dm1_test.shape
xfms = [np.eye(nv), xfm_procr, xfm_ridge, cha_procr, cha_ridge]
targets = dm2_test @ mapper
conn2 = 1 - cdist(targets.T, dm2_test.T, 'correlation')
iscs = []
for xfm in xfms:
aligned = dm1_test @ xfm
targets = aligned @ mapper
conn = 1 - cdist(targets.T, aligned.T, 'correlation')
isc = np.mean(zscore(conn, axis=0) * zscore(conn2, axis=0), axis=0)
iscs.append(isc)
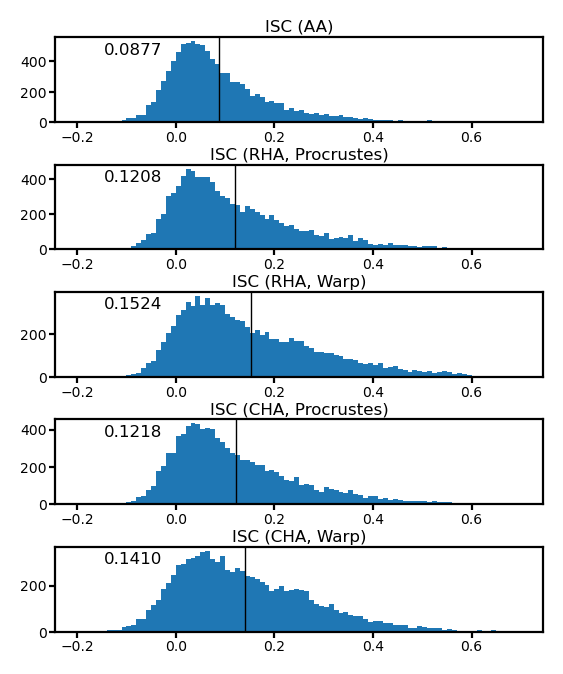
fig, axs = plt.subplots(5, 1, figsize=[_/2.54 for _ in [8, 10]], dpi=200)
for i, (ax, isc) in enumerate(zip(axs, iscs)):
ax.hist(isc, np.linspace(-0.4, 0.9, 131))
ax.axvline(isc.mean(), color='k', lw=0.5)
ax.tick_params('both', size=2, pad=1, labelsize=5)
title = ['ISC (AA)', 'ISC (RHA, Procrustes)', 'ISC (RHA, Warp)',
'ISC (CHA, Procrustes)', 'ISC (CHA, Warp)'][i]
ax.set_title(title, size=6, pad=2)
ax.annotate(f'{isc.mean():.4f}', (0.1, 0.8), xycoords='axes fraction', size=6)
fig.subplots_adjust(hspace=0.5)
plt.show()
nt, nv = dm1_test.shape
xfms = [np.eye(nv), xfm_procr, xfm_ridge, cha_procr, cha_ridge]
targets = dm2_test @ mapper
conn2 = 1 - cdist(targets.T, dm2_test.T, 'correlation')
cosines = []
for xfm in xfms:
aligned = dm1_test @ xfm
targets = aligned @ mapper
conn = 1 - cdist(targets.T, aligned.T, 'correlation')
norm = np.linalg.norm(conn, axis=0) * np.linalg.norm(conn2, axis=0)
cos = np.sum(conn * conn2 / norm, axis=0)
cosines.append(cos)
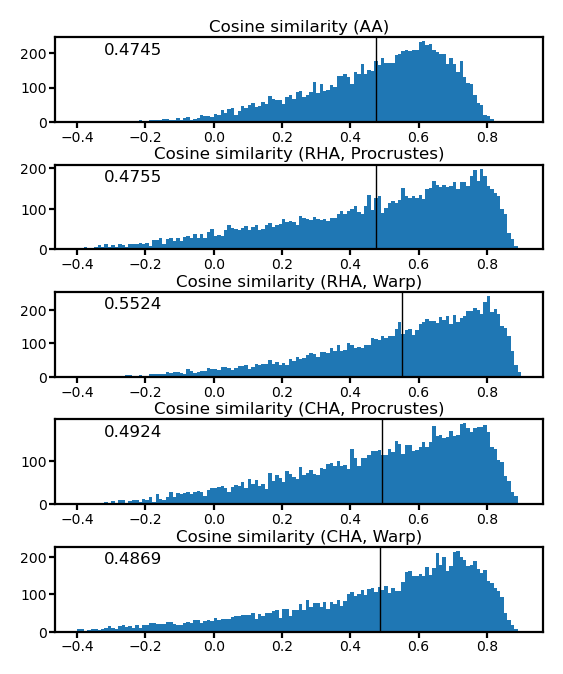
fig, axs = plt.subplots(5, 1, figsize=[_/2.54 for _ in [8, 10]], dpi=200)
for i, (ax, cos) in enumerate(zip(axs, cosines)):
ax.hist(cos, np.linspace(-0.4, 0.9, 131))
ax.axvline(cos.mean(), color='k', lw=0.5)
ax.tick_params('both', size=2, pad=1, labelsize=5)
title = 'Cosine similarity ' + ['(AA)', '(RHA, Procrustes)', '(RHA, Warp)',
'(CHA, Procrustes)', '(CHA, Warp)'][i]
ax.set_title(title, size=6, pad=2)
ax.annotate(f'{cos.mean():.4f}', (0.1, 0.8), xycoords='axes fraction', size=6)
fig.subplots_adjust(hspace=0.5)
plt.show()
Using zscored connectivity profiles#
cha_procr2 = searchlight_procrustes(
zscore(conn1, axis=0), zscore(conn2, axis=0), sls, dists, radius)
cha_ridge2 = searchlight_ridge(
zscore(conn1, axis=0), zscore(conn2, axis=0), sls, dists, radius)
pred_procr_cha2 = dm1_test @ cha_procr2
pred_ridge_cha2 = dm1_test @ cha_ridge2
isc_procr_cha2 = np.mean(
zscore(pred_procr_cha2, axis=0) * zscore(dm2_test, axis=0), axis=0)
isc_ridge_cha2 = np.mean(
zscore(pred_ridge_cha2, axis=0) * zscore(dm2_test, axis=0), axis=0)
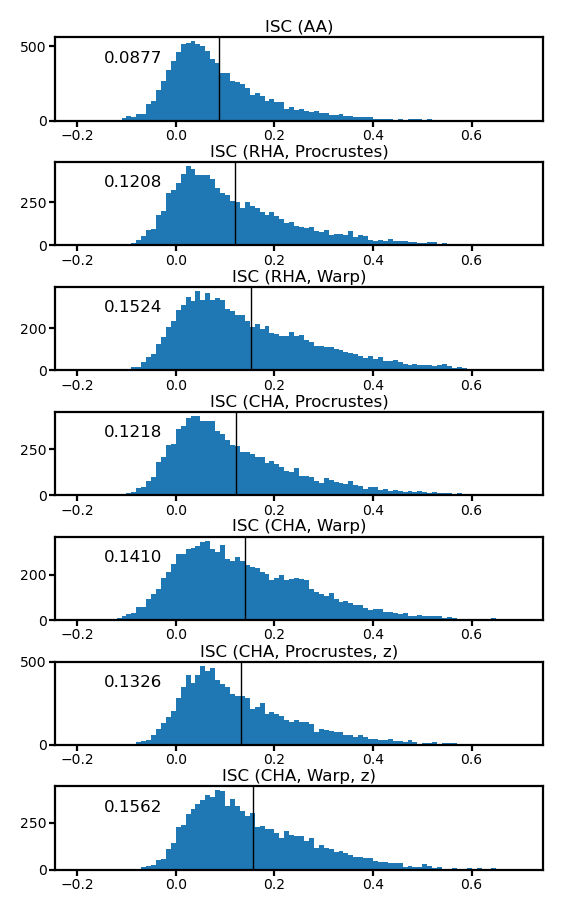
fig, axs = plt.subplots(7, 1, figsize=[_/2.54 for _ in [8, 14]], dpi=200)
iscs = [isc_orig, isc_procr, isc_ridge,
isc_procr_cha, isc_ridge_cha, isc_procr_cha2, isc_ridge_cha2]
for i, (ax, isc) in enumerate(zip(axs, iscs)):
ax.hist(isc, np.linspace(-0.2, 0.7, 91))
ax.axvline(isc.mean(), color='k', lw=0.5)
ax.tick_params('both', size=2, pad=1, labelsize=5)
title = ['ISC (AA)', 'ISC (RHA, Procrustes)', 'ISC (RHA, Warp)',
'ISC (CHA, Procrustes)', 'ISC (CHA, Warp)',
'ISC (CHA, Procrustes, z)', 'ISC (CHA, Warp, z)'][i]
ax.set_title(title, size=6, pad=2)
ax.annotate(f'{isc.mean():.4f}', (0.1, 0.7), xycoords='axes fraction', size=6)
fig.subplots_adjust(hspace=0.5)
plt.show()
nt, nv = dm1_test.shape
xfms = [np.eye(nv), xfm_procr, xfm_ridge, cha_procr, cha_ridge, cha_procr2, cha_ridge2]
targets = dm2_test @ mapper
conn2 = 1 - cdist(targets.T, dm2_test.T, 'correlation')
iscs = []
for xfm in xfms:
aligned = dm1_test @ xfm
targets = aligned @ mapper
conn = 1 - cdist(targets.T, aligned.T, 'correlation')
isc = np.mean(zscore(conn, axis=0) * zscore(conn2, axis=0), axis=0)
iscs.append(isc)
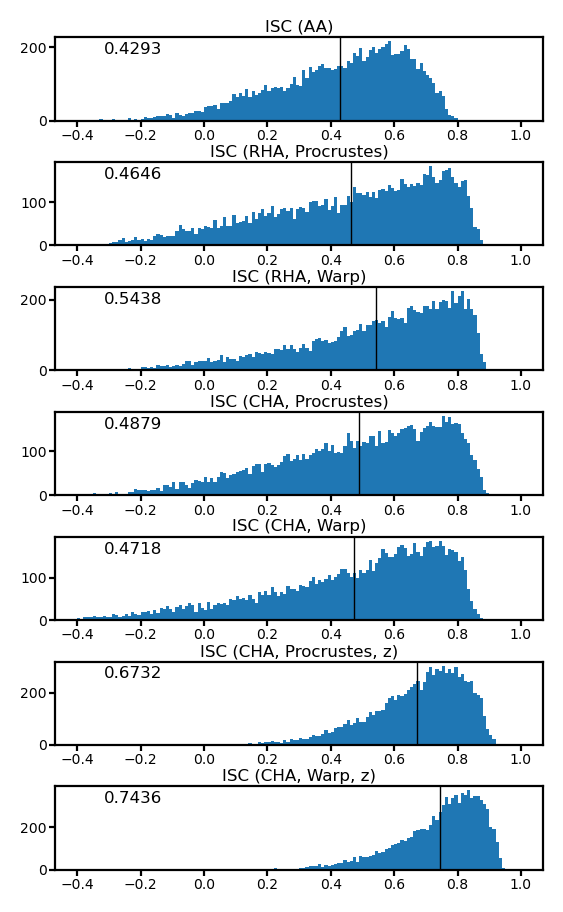
fig, axs = plt.subplots(7, 1, figsize=[_/2.54 for _ in [8, 14]], dpi=200)
for i, (ax, isc) in enumerate(zip(axs, iscs)):
ax.hist(isc, np.linspace(-0.4, 1.0, 141))
ax.axvline(isc.mean(), color='k', lw=0.5)
ax.tick_params('both', size=2, pad=1, labelsize=5)
title = ['ISC (AA)', 'ISC (RHA, Procrustes)', 'ISC (RHA, Warp)',
'ISC (CHA, Procrustes)', 'ISC (CHA, Warp)',
'ISC (CHA, Procrustes, z)', 'ISC (CHA, Warp, z)'][i]
ax.set_title(title, size=6, pad=2)
ax.annotate(f'{isc.mean():.4f}', (0.1, 0.8), xycoords='axes fraction', size=6)
fig.subplots_adjust(hspace=0.5)
plt.show()